How to extract a graph from your Python function
Published on Aug 5, 2024
10 min read
Prelude
You’re a Data Engineer. You write pipelines in Python which turn out to be quite complex. Maybe sometimes you take days - even weeks - off, just to come back and forget how everything is connected.
We talked before about data lineage and how useful it is to have an overview of the data flows in your team. It does not have to be a whole platform, such as Airflow or Dagster, where you spin up servers and the such. Let’s keep it simple; at least, as much as possible.
Let’s say you implemented a data pipeline. You have a Python function which does everything: fetch, process, and dump. Pretty standard. Orchestration, logging, metrics, etc. should be handled separately, such that one can look purely at the processing steps. Hmm, perhaps The Engineer should finally cover that topic too in a blogpost.
Anyway, so there’s a Python function, or simply a Python script that you trigger to do everything. Could we extract what that script does and pretty print a graph?
Give us an example!
From
def pipeline():
raw_data = read_data()
processed_data = process(raw_data)
write_data(processed_data)To
read_data -> data -> process_data -> processed -> write_dataBut with nicer formatting.
From nada to a graph
We have a couple of options here.
First, we could parse the codebase statically or at runtime (or even both ;)). The latter could be the only choice when the steps to execute are dynamically generated, e.g. a loop processing each file returned by an API.
Then, we need to decide how deep do we want to go. Can we assume that all the relevant steps are directly within this main function, or their internal calls are also relevant to see in the graph?
What about re-using variable names; df anyone?
What about calling the same function multiple times?
What about skipping some arbitrary things from the graph?
What about…
Ok, ok. It’s not so simple
It does not mean it’s impossible though. Let’s go step by step.
Decisions, decisions…
Let’s focus on doing things statically. Yes, this practically cuts off some possible scenarios from the start, but we need to start somewhere, right? It’s also arguably the easiest and the least invasive option.
Let’s also assume the codebase is nice and clean.

Nevertheless, we start from easy examples and build up. We look only at the contents of the main function; no recursion and so not going any deeper. We are not that brave to confront the Balrog.
So we want a simple function, such as
infer_graph(pipeline_func)that will show us an outline of what is happening in there.
Design
Perhaps we can also defer the pretty printing feature for now, and simply focus on extracting the relevant data. Practically, the nodes and the edges of the graph. One might already wonder though: which are the nodes here?
In the previous graph, it seemed like both the functions and the variables are nodes, while the edges have no associated metadata.
Why aren’t the variables nodes, while the functions edges?
Well, an argument here is about the first and the last steps of the graph, namely the first read and the last write. Do we want arrows pointing from or to nothing? Another argument are functions with multiple inputs and/or multiple outputs. Arrows in this case would make the graph rather confusing.
Hmm, but we then need to differentiate between nodes somehow, right?
Yes, though this mostly becomes a problem for the presentation side of things, which we gracefully already deferred :)
For those having déjà vu, recall for example the Marquez UI over OpenLineage events or the Dagster concept of an @asset.
Code
OK, finally some code.
We are taking an iterative approach here until we reach a relatively decent v0 version of our infer_graph function.
In terms of dependencies, we actually only need a core library import: ast.
Recall the pipeline we want to infer a graph from, now with more details but still very simplistic.
def read_data(*args, **kwargs) -> None:
return 1
def process(data: int) -> int:
return data + 1
def write_data(data: int) -> None:
print(data)
def pipeline():
raw_data = read_data()
processed_data = process(raw_data)
write_data(processed_data)Now I am by no means an expert in such meta-programming. I am sure those well versed in the intricacies of Python will easily spot edge cases or better approaches. If you happen to be one of them, I encourage you to share feedback in the Github repo I will share towards the end :)
The most basic approach here is the following.
We statically inspect the main function and only look at its direct children, i.e. the contents of this function and no further.
We only look for Assign and Expr statements.
Given our pipeline function, these should cover all the nodes.
This looks like:
import ast
import inspect
def infer_graph(func):
entire_ast = ast.parse(inspect.getsource(func))
entire_pipeline_func = entire_ast.body[0]
edges = list()
for elem in entire_pipeline_func.body:
if isinstance(elem, ast.Assign):
# e.g. given: processed_data = process(data)
output_ = elem.targets[0].id # processed_data
f = elem.value.func.id # process
input_ = elem.value.args[0].id if elem.value.args else None # data
edges.append((
input_, f, output_
))
elif isinstance(elem, ast.Expr):
f = elem.value.func.id
input_ = elem.value.args[0].id if elem.value.args else None
edges.append((
input_, f, None
))
return edgesThe Expr statement means we call a function without any output, therefore we can assume this is a terminal node in the graph.
An example here is the write_data function call.
The Assign statement means we assign something else to a variable, such as storing the output of read_data into raw_data.
Here we assume that all variables should be part of the graph; we’ll get to ignoring the irrelevant nodes soon enough.
Since when assigning you take an input and store an output, we already know that the input is where an arrow would point from and the output is where it would point to.
At the moment, we assume that each input in these assignments is actually a function processing something from another variable passed as an argument.
This means that the just mentioned input is in reality 2 nodes: the input data, and the processing function.
So taking the example of processed_data = process(data) we extract 3 nodes:
dataas the input data node,processas the function node, andprocessed_dataas the output data node.
To use our function, we
print(infer_graph(pipeline))And the crude output looks like
[(None, 'read_data', 'raw_data'), ('raw_data', 'process', 'processed_data'), ('processed_data', 'write_data', None)]It is easy to see how read_data is then the first node of our graph, creating the raw_data node, which gets picked up by process, and so on.
So what now?
What we have so far is a veeeeery simplistic function. Let’s try to tackle some of the scenarios we already pointed out. First, we’ll need a slightly more complex pipeline.
Note that from this point onwards, only pseudo-diffs will be shown else this blogpost will explode in size.
- def pipeline():
- raw_data = read_data()
+ def pipeline(not_a_node_arg: int):
+ raw_data = read_data(not_a_node_arg)
+ not_a_node_variable = not_a_node_arg
...
+ print(not_a_node_arg)We assign a constant to a variable, therefore we no longer assume every Assign has a function on its right.
So let’s update our infer_graph to only look at functions, here Call statements.
- if isinstance(elem, ast.Assign):
+ if isinstance(elem, ast.Assign) and isinstance(elem.value, ast.Call):Which produces the output:
[('not_a_node_arg', 'read_data', 'raw_data'), ('raw_data', 'process', 'processed_data'), ('processed_data', 'write_data', None), ('not_a_node_arg', 'print', None)]Well, we correctly ignore the assignment when the right side is not a function. Again, we assume that data must be processed through a function to “produce” a new node.
However the output still does not look quite right.
We have the irrelevant not_a_node_arg being shown, plus a function using that and only that arg.
We should not show those.
We assume the entire graph is created within the main function we infer a graph from.
The main change is to keep track of the nodes - more specifically the “data” variables - seen so far, adding them when encountering an Assign.
Then when encountering an Expr - so a function call with no assignment - to only add the edge when the input is from a known node.
+ nodes = set()
...
+ nodes.add(output_)
...
+ if input_:
edges.append((
...Lastly, we need to filter function arguments for those known nodes.
- input_ = elem.value.args[0].id if elem.value.args else None
+ input_ = elem.value.args[0].id if elem.value.args and elem.value.args[0].id in nodes else NoneWith these updates, we again have the expected output of
[(None, 'read_data', 'raw_data'), ('raw_data', 'process', 'processed_data'), ('processed_data', 'write_data', None)]But this is still too simplistic. A real life pipeline is much more complex.
Let’s complicate it further
We need a third version of the pipeline:
- raw_data = read_data(not_a_node_arg)
+ raw_data = read_data(not_a_node_arg, not_a_node_kwarg=42)
...
- processed_data = process(raw_data)
+ processed_data = process(data=raw_data)We now use multiple args and kwargs. Some of them can be different to a variable name.
Parsing args is now definitely more complicated.
We need to use an iterable data structure - such as a list - to store the inputs while checking for the expected ast types; and we do this for both args and kwargs.
Finally to generalize a bit, we extract this into a separate function.
+ def extract_input(elem, nodes) -> List[str]:
+ args = list()
+ for arg in elem.value.args:
+ if isinstance(arg, ast.Name):
+ args.append(arg.id)
+ elif isinstance(arg, ast.Constant):
+ args.append(arg.value)
+ kwargs = list()
+ for kwarg in elem.value.keywords:
+ if isinstance(kwarg, ast.Name):
+ kwargs.append(kwarg.value.id)
+ elif isinstance(kwarg, ast.keyword) and isinstance(kwarg.value, ast.Name):
+ kwargs.append(kwarg.value.id)
+ inputs = [input_ for input_ in args + kwargs if input_ in nodes]
+
+ return inputsWhich we use within our infer_graph.
- input_ = elem.value.args[0].id if elem.value.args and elem.value.args[0].id in nodes else None # data
+ input_ = extract_input(elem, nodes) # [data]And we still have the expected output, with the change that each “edge” can now have multiple inputs.
[([], 'read_data', 'raw_data'), (['raw_data'], 'process', 'processed_data'), (['processed_data'], 'write_data', None)]So we are done now?
Yes :)

The code part is over. Given our assumptions:
- a single main function with all our processing steps,
- purely processing steps, so no logging, profiling, etc.,
- no recursive checks within the main function.
We implemented a function infer_graph which statically extracts a graph from our main data processing pipeline function.
Success!
Well not really. Our function is not bullet proof. There are still situations where it would under-perform. To name a few:
- what if we have multiple outputs from a function?
- what if we re-use variable names?
- what if function arguments have different types? lambdas, other functions calls, etc.?
- what about classes?
- can you think of others?
We could also challenge our starting assumptions:
- what if we actually have multiple main functions that should be part of the same graph?
- what if we want to ignore certain functions?
- what if we want to go deeper within the functions?
Some alternatives
Visitor pattern
Those familiar with the ast package can finally take a deep breath; yes, one could implement the visitor pattern.
The structure is pretty simple, however we still need clever filtering and tracking steps.
One needs to implement visit_ methods while keeping track of the parents and updating the graph where relevant.
So we still need to implement the same kind of checks we did in this blogpost! Perhaps more tricky, however this way one could traverse the function as deep as desired.
Debugger… for data?
Another option - or perhaps additionally - is to build the graph at runtime. This would finally enable those dynamic nodes we previously mentioned. Also one could time the steps for a win-win!
To do this, one needs a machinery fairly similar to a debugger and as it turns out, Python 3.12 makes it that much easier with the sys.monitoring package. Essentially, each time Python ends up executing something, say, a function, we can register a callback. This callback could then build the graph for us!
Nevertheless, we still struggle with the same challenges: we don’t care about everything happening during execution. We only care about some specific functions for data processing and how they are connected.
Addendum
Didn’t you say this code is posted somewhere? We have feedback!
Yes, it is posted here. Do start a discussion!
Is creating this graph really worth the effort?
For such a small pipeline, definitely not. The goal is to have these for complex projects, and more specifically, those with many many pipelines. Nobody can remember them all! Do check the previous blogpost on data lineage in general.
Why did you skip the presentation layer of the graph? It’s the most important!
Well here’s an example achievable via networkx with matplotlib.
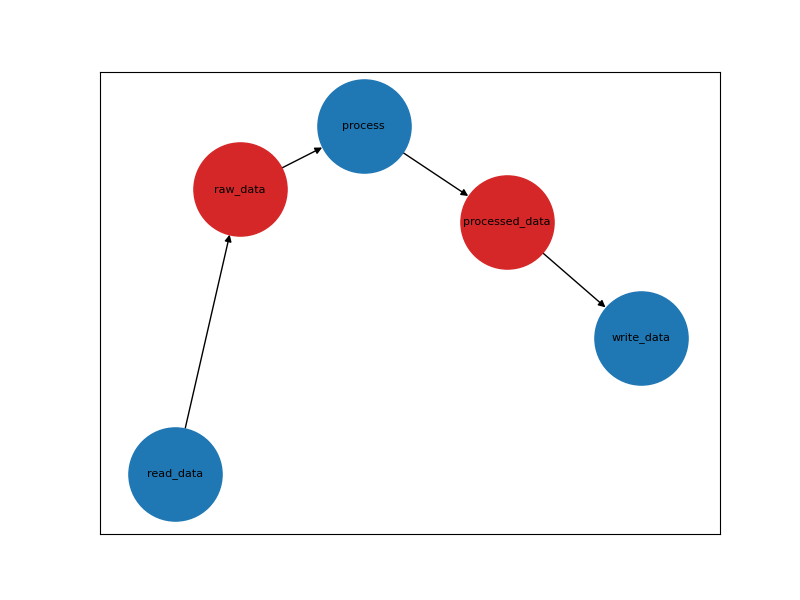
So it is doable but making it visually appealing is a whole separate topic.